Note
Click here to download the full example code
Fitting Distributions¶
Histograms, PDF fits, Kernel Density.
from __future__ import absolute_import, print_function, division
# Author: Moritz Lotze <mlotze@km3net.de>
# License: BSD-3
import matplotlib.pyplot as plt
import numpy as np
import statsmodels.api as sm
from scipy.stats import norm
from sklearn.mixture import GaussianMixture
from sklearn.model_selection import GridSearchCV
from sklearn.neighbors import KernelDensity
import km3pipe.style.moritz # noqa
First generate some pseudodata: A bimodal gaussian, + noise.
N = 100
bmg = np.concatenate((
np.random.normal(15, 1, int(0.3 * N)),
np.random.normal(20, 1, int(0.7 * N))
))
noise_bmg = 0.5
data = np.random.normal(bmg, noise_bmg)[:, np.newaxis]
# make X axis for plots
x = np.linspace(5, 35, 3 * N + 1)
Histograms (nonparametric)¶
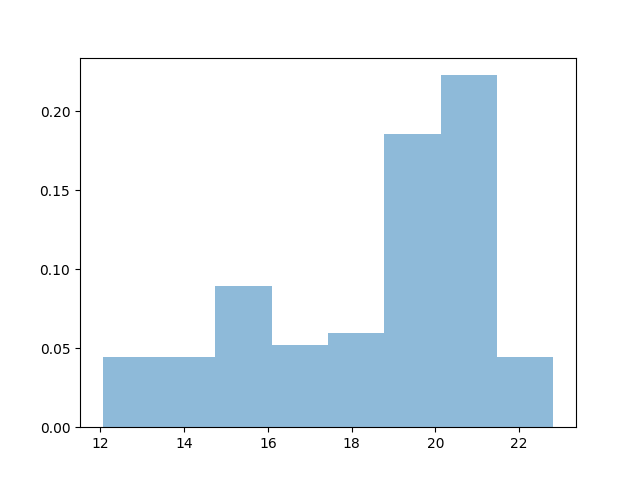
The simplest nonparametric density estimation tool is the Histogram. Choosing the binning manually can be tedious, however:
15 bins, spaced from data.min() to data.max().
plt.hist(data, bins=15, alpha=.5, normed=True)
Fit Distribution via Maximum Likelihood¶
If we have a hypothesis what the distribution looks like (e.g. gaussian), and want to fit its parameters.
The nice thing is, you can define your own PDFs in scipy and fit it. Or take one from the dozens of pre-defined ones.
However, there is no bimodal gaussian implemented in scipy yet :/ In this case, either define it yourself, or use a GMM (below)
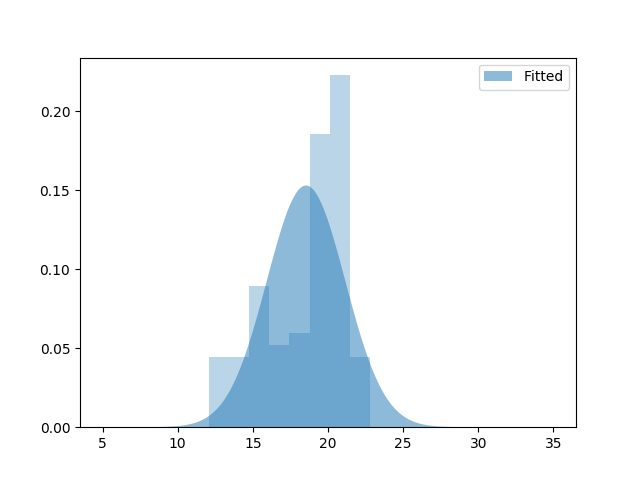
mu, sig = norm.fit(data)
plt.fill_between(x, norm(mu, sig).pdf(x), alpha=.5, label='Fitted')
plt.legend()
print('Unimodal Gaussian Fit: Mean {:.4}, stdev {:.4}'.format(mu, sig))
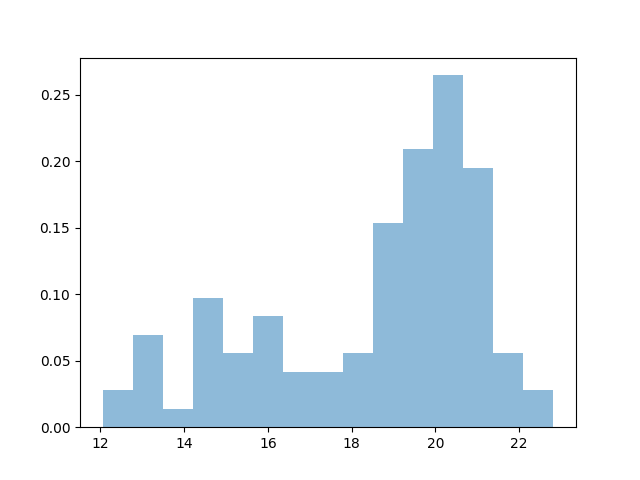
plt.hist(data, bins='auto', alpha=.3, normed=True)
Out:
Unimodal Gaussian Fit: Mean 18.54, stdev 2.608
As expected, the result is rather silly, since we are only fitting one of the two gaussians.
Fit Gaussian Mixture Model (GMM)¶
Assuming the data is the sum of one or more gaussians. Easily handles multidimensional case as well.
gmm = GaussianMixture(n_components=2, covariance_type='spherical')
gmm.fit(data)
mu1 = gmm.means_[0, 0]
mu2 = gmm.means_[1, 0]
var1, var2 = gmm.covariances_
wgt1, wgt2 = gmm.weights_
print(
'''Fit:
1: Mean {:.4}, var {:.4}, weight {:.4}
2: Mean {:.4}, var {:.4}, weight {:.4}
'''.format(mu1, var1, wgt1, mu2, var2, wgt2)
)
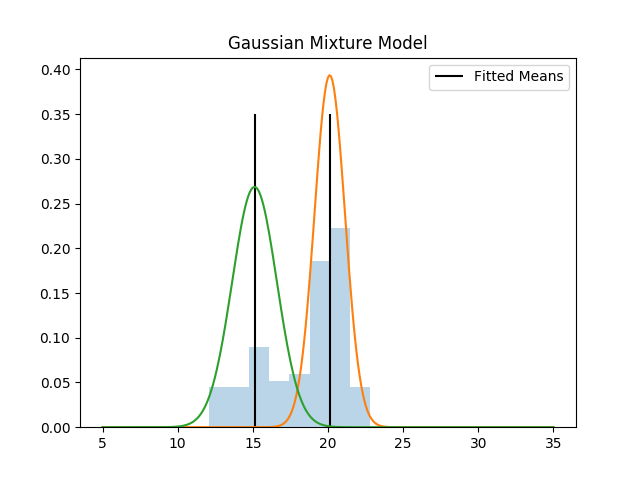
plt.hist(data, bins='auto', alpha=.3, normed=True)
plt.vlines((mu1, mu2), ymin=0, ymax=0.35, label='Fitted Means')
plt.plot(x, norm.pdf(x, mu1, np.sqrt(var1)))
plt.plot(x, norm.pdf(x, mu2, np.sqrt(var2)))
plt.legend()
plt.title('Gaussian Mixture Model')
Out:
Fit:
1: Mean 20.12, var 1.028, weight 0.685
2: Mean 15.11, var 2.205, weight 0.315
Kernel Density: (non-parametric)¶
If we have no strong assumptions about the underlying pdf.
“Smooth out” each event with a kernel (e.g. gaussian) of a certain bandwidth, then add together all these mini-functions.
The “bandwidth” (width of the kernel function) depends on the data, and can be estimated using cross-validation + maximum likelihood
in Statsmodels
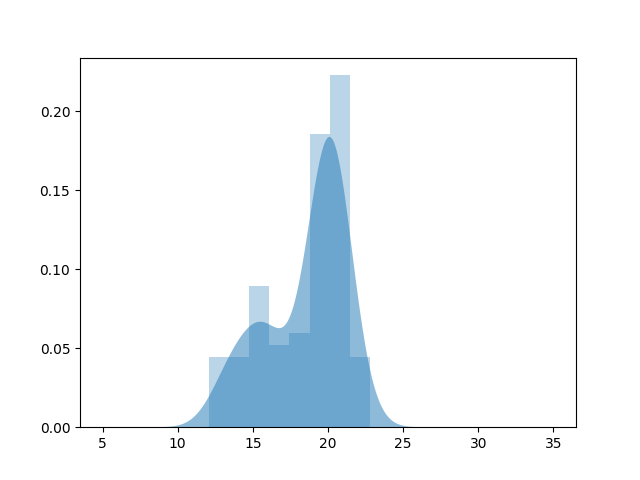
dens = sm.nonparametric.KDEUnivariate(data)
dens.fit()
kde_sm = dens.evaluate(x)
plt.fill_between(x, kde_sm, alpha=.5, label='KDE')
plt.hist(data, bins='auto', alpha=.3, normed=True)
in scikit-learn
params = {'bandwidth': np.logspace(-2, 2, 50)}
grid = GridSearchCV(KernelDensity(), params)
grid.fit(data)
print("best bandwidth: {0}".format(grid.best_estimator_.bandwidth))
# use the best estimator to compute the kernel density estimate
kde_best = grid.best_estimator_
kde_sk = np.exp(kde_best.score_samples(x[:, np.newaxis]))
plt.fill_between(x, kde_sk, alpha=.5, label='KDE')
plt.hist(data, bins='auto', alpha=.3, normed=True)
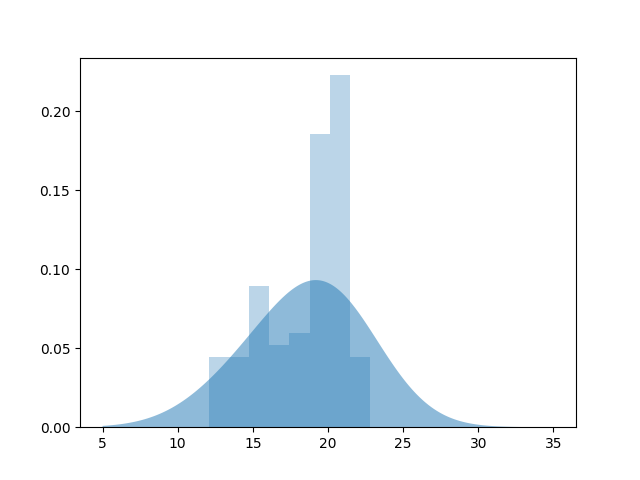
Out:
best bandwidth: 3.3932217718953264
References¶
- B.W. Silverman, “Density Estimation for Statistics and Data Analysis”
- Hastie, Tibshirani and Friedman, “The Elements of Statistical Learning: Data Mining, Inference, and Prediction”, Springer (2009)
- Liu, R., Yang, L. “Kernel estimation of multivariate cumulative distribution function.” Journal of Nonparametric Statistics (2008)
Total running time of the script: ( 0 minutes 3.621 seconds)